Format Specification
Overview
The Spacewalk binary file format (.sw) uses HDF5 to store and organize genomic spatial data. The format supports both single-point (centroid) and multi-point (cluster) data types.
File Structure
Header Group
Required top-level group named header with metadata:
| Key | Value |
|---|---|
| format | “sw” |
| genome | A valid genimd id. e.g., “hg19” |
| pointtype | “SINGLE_POINT” or “MULTI_POINT” |
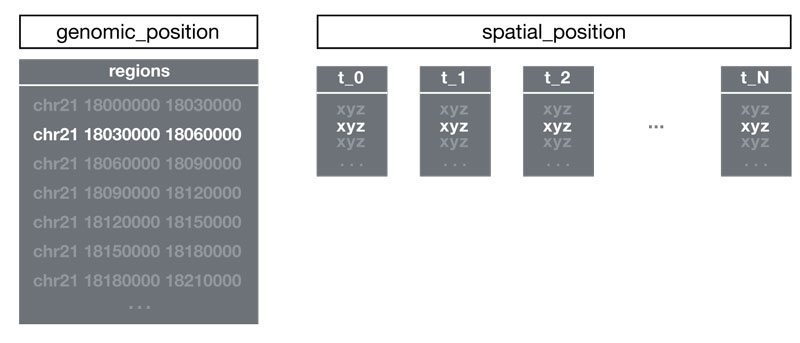
Genomic Position
The genomic_position group contains:
- regions dataset: 2D array with columns:
- chromosome name (string)
- region start (integer, bp)
- region end (integer, bp)
Regions must be sorted by genomic start position.
Spatial Position
The spatial_position group contains XYZ data in two possible formats:
Single Point Format
- One XYZ point per genomic region
- Datasets named
t_0,t_1, etc. for each trace - Direct 1:1 mapping between regions and points
Multi Point Format
- Multiple XYZ points per genomic region
- Trace groups named
t_0,t_1, etc. - Each row format:
region_index x y z - Flexible mapping between regions and points
Example Files
View example files in HDF5 format: