Legacy Spacewalk Text Format
The legacy text-based format (.swt) is now deprecated. Use the swt2sw tool to migrate to the new .sw format.
Format Specification
File Structure
- First Line: Format directive with key-value properties
- Format specifier (required)
- Name (required)
- Genome identifier (required, e.g., hg38, GRCh38)
-
Second Line: Six column headings, whitespace-delimited
- Remaining Lines: Blocks of trace data
- Each block begins with a “trace” line
- Followed by data lines
- All tokens are whitespace delimited
Example Formats
Ball & Stick Data (ORCA)
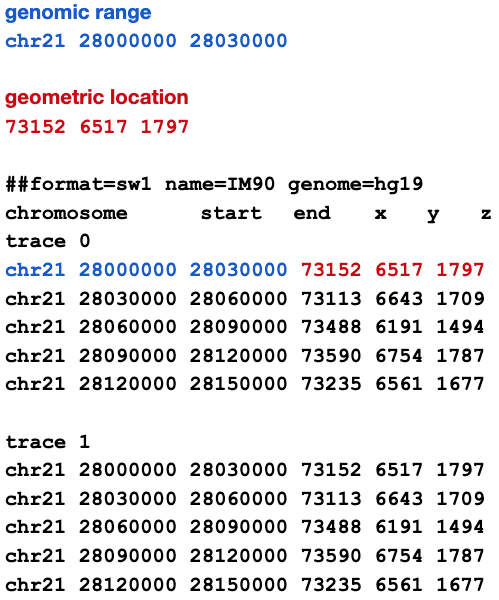
Each record links a geometric location with a genomic range:
Point Cloud Data (OligoSTORM)
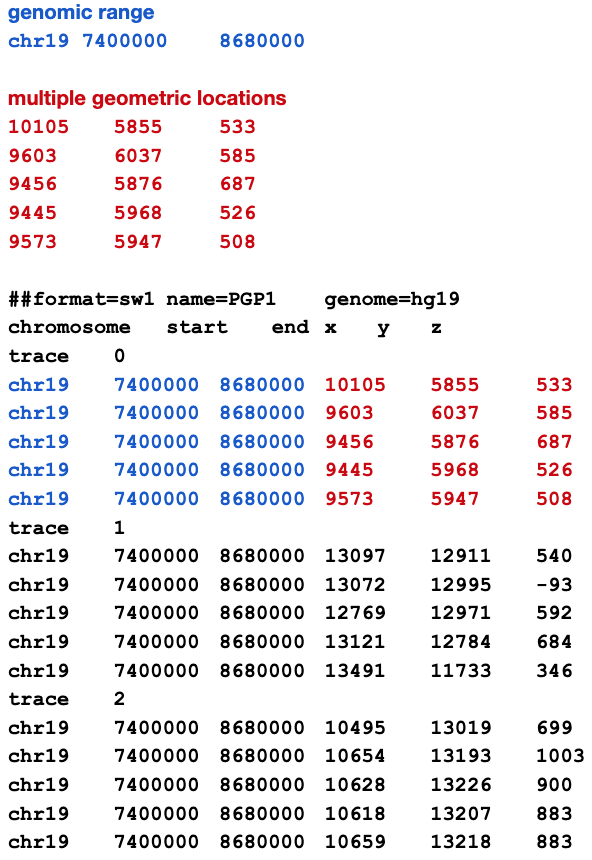
Multiple geometric locations can share the same genomic range:
Migration
To convert legacy .swt files to the new .sw format:
- Install the migration tool:
pip install swt2sw - Convert your files:
swt2sw input.swt output.sw