File Structure and Organization
The Spacewalk binary file format is based on the Hierarchical Data Format HDF5.
HDF5 Concepts
Key HDF5 concepts used in the .sw format are:
- Group: The HDF5 analog to a directory or folder of a file system
- Dataset: An M by N array that stores a particular chunk data in the file
- Attributes: A property of groups and datasets that enables optional storage of associated metadata as key/value pairs
Since the .sw format is a valid HDF5 file, the contents can be examined by any general HDF5 viewer, such as myHDF5.
File Organization
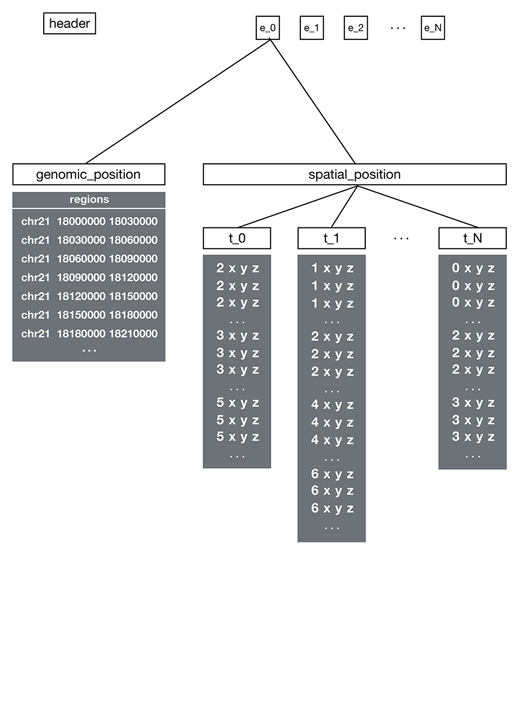
The file contains:
1. Header Group
Must be named header with required metadata:
| Key | Value |
|---|---|
| format | “sw” |
| genome | A valid genimd id. e.g., “hg19” |
| pointtype | “SINGLE_POINT” or “MULTI_POINT” |
2. Ensemble Groups
Each ensemble group contains data for a single experiment with:
- Unique names (except “header”)
- Optional “name” attribute
- Two child groups:
genomic_positionandspatial_position
Data Types
Single Point Data
- One XYZ point per genomic region
- Used for centroid-based visualization
- Supports ball-and-stick rendering
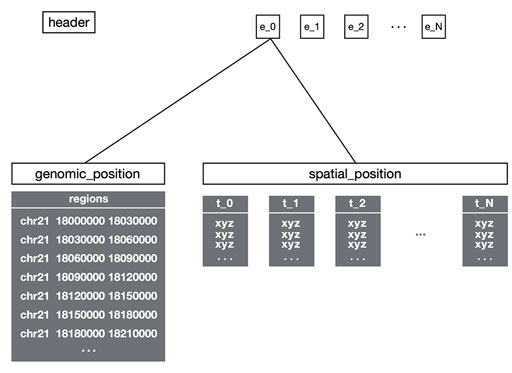
Multi Point Data
- Multiple XYZ points per genomic region
- Used for point cloud visualization
- Supports density-based rendering